Computational Geometry,Fundamental Structures:Introduction and Arrangements
Introduction
Computational geometry deals with the design and analysis of algorithms and data structures for problems involving spatial data. The questions that are studied range from basic problems such as line-segment intersection (“Compute all intersection points in a given set of line segments in the plane.”) to quite involved problems such as motion-planning (“Compute a collision-free path for a robot in workspace from a given start position to a given goal position.”) Because spatial data plays an important role in many areas within and outside of computer science—CAD/CAM, computer graphics and virtual reality, and geography are just a few examples—computational geometry has a broad range of applications. Computational geometry emerged from the general algorithms area in the late 1970s. It experienced a rapid growth in the 1980s and by now is a recognized discipline with its own conferences and journals and many researchers working in the area. It is a beautiful field with connections to other areas of algorithms research, to application areas like the ones mentioned earlier, and to areas of mathematics such as combinatorial geometry.
To design an efficient geometric algorithm or data structure, one usually needs two ingredients: a toolbox of algorithmic techniques and geometric data structures and a thorough understanding of the geometric properties of the problem at hand. As an example, con- sider the classic post-office problem, where we want to preprocess a set S of n points in the plane—the points in S are usually called sites —for the following queries: report the site in S that is closest to a query point q. A possible approach is to subdivide the plane into n regions, one for each site, such that the region of a site s ∈ S consists of exactly those points q ∈ R2 for which s is the closest site. This subdivision is called the Voronoi diagram of S. A query with a point q can now be answered by locating the region in which q lies, and reporting the site defining that region. To make this idea work, one needs an efficient data structure for point location. But one also needs to understand the geometry: What does the Voronoi diagram look like? What is its complexity? How can we construct it efficiently?
In Part IV, many data structures for spatial data were already discussed.
Hence, in this chapter we will focus on the second ingredient: we will discuss a number of basic geometric concepts. In particular, we will discuss arrangements in Section 62.2, convex hulls in Section 62.3, Voronoi diagrams in Section 62.4, and triangulations in Section 62.5.
More information on computational geometry can be found in various sources: there are several general textbooks on computational geometry [7, 8, 45, 49], as well as more special- ized books e.g. on arrangements [20, 53] and Voronoi diagrams [44]. Finally, there are two handbooks that are devoted solely to (discrete and) computational geometry [24, 50].
Arrangements
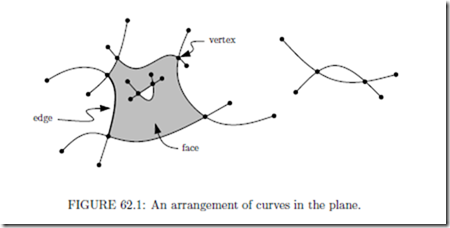
The arrangement A(S) defined by a finite collection S of curves in the plane is the sub-division of the plane into open cells of dimensions 2 (the faces), 1 (the edges ), and 0 (the
higher dimensions: the arrangement defined by a set S of geometric objects in Rd such as hyperplanes or surfaces, is the decomposition of Rd into open cells of dimensions 0,... ,d induced by S. The cells of dimension k are usually called k-cells. The 0-cells are called vertices, the 1-cells are called edges, the 2-cells are called faces, the (d − 1)-cells are called facets, and the d-cells are sometimes just called cells.
Arrangements have turned out to form a fundamental concept underlying many geometric problems and the efficiency of geometric algorithms is often closely related to the combinatorial complexity of (certain parts of) some arrangement. Moreover, to solve a certain problem geometric algorithms often rely on some decomposition of the arrangement underlying the problem. Hence, the following subsections give some more information on the complexity and the decomposition of arrangements.
Let H be a collection of n hyperplanes in Rd. As stated earlier, the arrangement A(H) is the decomposition of Rd into open cells of dimensions 0,... ,d induced by H. The combinatorial complexity of A(H) is defined to be the total number of cells of the various dimensions.
This definition immediately carries over to arrangements induced by other objects, such as segments in the plane, or surfaces in Rd. For example, the complexity of the arrangement in Fig. 62.1 is 58, since it consists of 27 vertices, 27 edges, and 4 faces (one of which is the unbounded face).
It is easy to see that the maximum complexity of an arrangement of n lines in the plane is Θ(n2): there can be at most n(n − 1)/2 vertices, at most n2 edges, and at most n2/2+ n/2 + 1 faces. Also for an arrangement of curves the maximum complexity is Θ(n2), provided that any pair of curves intersects at most s times, for a constant s. More generally, the maximum complexity of an arrangement of n hyperplanes in Rd is Θ(nd). The same bound holds for well-behaved surfaces (such as algebraic surfaces of constant maximum degree) or well-behaved surface patches in Rd.
Single cells. It becomes more challenging to bound the complexity when we consider only a part of an arrangement. For example, what is the maximum complexity of a single cell, that is, the maximum number of i-cells, for i < d, on the boundary of any given d-cell? For lines in the plane this is still rather easy—the maximum complexity is Θ(n), since any line can contribute at most one edge to a given face—but the question is already quite hard for arrangements of line segments in the plane. Here it turns out that the maximum complexity can be Θ(nα(n)), where α(n) is the extremely slowly growing functional inverse of Ackermann’s function. More generally, for Jordan curves where each pair intersects in at most s points, the maximum complexity is Θ(λs+2(n)), where λs+2(n) is the maximum length of a Davenport-Schinzel sequence of order s+2 on n symbols. The function λs+2(n) is only slightly super-linear for any constant s. In higher dimensions, tight bounds are known for hyperplanes: the famous Upper Bound Theorem states that the maximum complexity of a single cell is Θ(nLd/2J). For most other objects, the known upper and lower bounds are close but not tight—see Table 62.1.
Lower envelopes. Another important substructure is the lower envelope. Intuitively, the
lower envelope of a set of segments in the plane is what one would see when looking at the segments from below—see Fig. 62.2. More formally, if we view the segments as graphs of partially defined (linear) functions, then the lower envelope is the point-wise minimum of these functions. Similarly, the upper envelope is defined as the point-wise maximum of the functions. The definition readily extends to x-monotone curves in the plane, to planes, triangles, or xy-monotone surface patches in R3, etc.
Envelopes are closely related to single cells. The lower envelope of a set of lines in the plane, for instance, is the boundary of the single cell in the arrangement that is below all the lines. The vertices of the lower envelope of a set of segments in the plane are also vertices of the unbounded cell defined by those segments, but here the reverse is not true: vertices of the unbounded cell that are above other segments are not on the lower envelope. Nevertheless, the worst-case complexities of lower envelopes and single cells are usually very similar—see Table 62.1.
Other substructures. More types of substructures have been studied than single cells and envelopes: zones, levels, multiple cells, etc. The interested reader may consult the Chapter 21 of the CRC Handbook of Discrete and Computational Geometry books by Edelsbrunner [20] or Sharir and Agarwal [53].
Decomposition
Full arrangements, or substructures in arrangements, are by themselves not convenient to work with, because their cells can be quite complex. Thus it is useful to further decompose the cells of interest into constant-complexity subcells: triangles or trapezoids in 2D, and simplices or trapezoid-like cells in higher dimensions. There are several ways of doing this.
Bottom-vertex triangulations. For arrangements of hyperplanes, the so-called bottom- vertex triangulation is often used. This decomposition is obtained as follows. Consider a bounded face f in a planar arrangement of lines. We can decompose f into triangles by drawing a line segment from the bottommost vertex v of f to all other vertices of f , except the vertices that are already adjacent to v—see Fig. 62.3(a). Note that this easy method for triangulating f is applicable since f is always convex. To decompose the whole arrangement of lines (or some substructure in it) we simply decompose each face in this manner.∗
To decompose a d-cell C in a higher-dimensional arrangement of hyperplanes, we proceed inductively as follows. We first decompose each (d − 1)-cell on the boundary of C, and then extend each (d − 1)-simplex in this boundary decomposition into a d-simplex by connecting
∗Unbounded faces require a bit of care, but they can be handled in a similar way.
each of its vertices to the bottommost vertex of C. Again, this can readily be used to decompose any subset of cells in an arrangement of hyperplanes.
The total number of simplices in a bottom-vertex decomposition is linear in the total complexity of the cells being decomposed.
Vertical decompositions. The bottom-vertex triangulation requires the cells to be con- vex, so it does not work for arrangements of segments or for arrangements of surfaces. For such arrangements one often uses the vertical decomposition (or: trapezoidal decomposition). For an arrangement of line segments or curves in the plane, this decomposition is defined as follows. Each vertex of the arrangement—this can be a segment endpoint or an intersection point—has a vertical connection to the segment immediately above it, and to the segment immediately below it. If there is no segment below or above a vertex, then the connection extends to infinity. This decomposes each cell into trapezoids: subcells that are bounded by at most two vertical connections and by at most two segments—see Fig. 62.3(b).
This definition can be generalized to higher dimensions as follows. Suppose we wish to decom- pose the arrangement A(S) induced by a collection S of surfaces in Rd, where each surface is vertically monotone (that is, any line parallel to the xd-axis intersects the surface in at most one point). Each point of any (d − 2)-dimensional cell of A(S) is connected by a vertical segment to the surface immediately above it and to the surface immediately below it. In other words, from each (d − 2)-cell we extend a vertical wall upward and downward.
These walls together decompose the cells into subcells bounded by vertical walls and by at most two surfaces from S—one from above and one from below. These subcells are vertically monotone, but do not yet have constant complexity. Hence, we recursively decompose the bottom of the cell, and then extend this decomposition vertically upward to obtain a decomposition of the entire cell.
The vertical decomposition can be used for most arrangements (or substructures in them). In the plane, the maximum complexity of the vertical decomposition is linear in the total complexity of the decomposed cells. However, in higher dimensions this is no longer true. In R3, for instance, the vertical decomposition of an arrangement of n disjoint triangles can consist of Θ(n2) subcells, even though in this case the total complexity of the arrangement in obviously linear. Unfortunately, this is unavoidable, as there are collections of disjoint triangles in R3 for which any decomposition into convex subcells must have Ω(n2) subcells. For n intersecting triangles, the vertical decomposition has complexity O(n2α(n) log n+ K), where K is the complexity of the arrangement of triangles [56]. More information about the complexity of vertical decompositions in various settings can be found in Halperin’ssurvey on arrangements [24, Chapter 21].
In many cases, vertical decompositions can be constructed in time proportional to their complexity, with perhaps a small (logarithmic or O(nε)) multiplicative factor.
Duality Consider the transformation in the plane that maps the point p = (px, py ) to the line p∗ : y = pxx−py , and the line £ : y = ax+b to the point £∗ = (a, −b). Such a transformation that maps points to lines and vice versa is called a duality transform. Often the term primal plane is used for the plane in which the original objects live, and the term dual plane is used for the plane in which their images live. The duality transform defined above has a few easy-to-verify properties:
(i) It is incidence preserving: if a point p lies on a line £, then the point £∗ dual to £ lies on the line p∗ dual to p.
(ii) It is order preserving: if a point p lies above a line £, then £∗ lies above p∗.
These properties imply several others. For example, three points on a line become three lines through a point under the duality transform—see Fig. 62.4. Another property is that for any point p we have (p∗)∗ = p. Notice that the duality transform above is not defined for vertical lines. This technicality is usually not a problem, as vertical lines can often be handled separately.
This duality transform is so simple that it does not seem very interesting at first sight. However, it turns out to be extremely useful. As an example, consider the following problem. We are given a set P of n points in the plane, which we wish to preprocess for strip-emptiness queries: given a query strip—a strip is the region between two parallel lines— decide whether that strip is empty or if it contains one or more points from P . If we know about duality, and we know about data structures for point location, then this problem is easy to solve:
we take the duals of the points in P to obtain a set P ∗ of lines in the plane, and we preprocess the arrangement A(P ∗) induced by P ∗ for logarithmic-time point location. To decide whether the strip bounded by lines £1 and £2 is empty, we perform point locations with £∗ and £∗ in A(P ∗); the strip is empty if and only if £∗ and £∗ lie in the same face of the arrangement. (This does not work if £1 and £2 are vertical, since then their duals are undefined. But for this case we can simply build one extra data structure, which is a balanced binary search tree on the x-coordinates of the points.)
In principle, of course, we could also have arrived at this solution without using duality. After all, duality does not add any new information: it is just a different way of looking at things. Hence, every algorithm or data structure that works in the dual plane can also be interpreted as working in the primal plane. But some things are simply more easy to see in the dual plane. For example, a face in the arrangement A(P ∗) in the dual plane is much more visible than the collection of all lines in the primal plane dividing P into two subsets in a certain way. So without duality, we would not have realized that we could solve the strip-emptiness problem with a known data structure, and we would probably not have been able to develop that structure ourselves either.
This is just one example of the use of duality. There are many more problems where duality is quite useful. In fact, we will see another example in the next section, when we study convex hulls.






Comments
Post a Comment